California Conservation Genomics Project
 In 2019, we took the leading role in securing $12M in funding for a major new initiative, the California Conservation Genomics Project (CCGP). The project includes all 10 UC campuses, about 100 PIs and co-PIs, and now covers 135 projects, and 240 species of plants and animals, including marine, freshwater, and terrestrial taxa. In addition to administering the entire CCGP, the Shaffer Lab is collaborating with Ian Wang’s group at UC Berkeley and multiple state and federal agency scientists to produce landscape genomic data and analyses for 34 species of reptiles and amphibians covering the breadth of California. Our goal is to produce a comprehensive analysis of the geography of genetic variation to guide management and habitat protection now and in the face of climate change.
In 2019, we took the leading role in securing $12M in funding for a major new initiative, the California Conservation Genomics Project (CCGP). The project includes all 10 UC campuses, about 100 PIs and co-PIs, and now covers 135 projects, and 240 species of plants and animals, including marine, freshwater, and terrestrial taxa. In addition to administering the entire CCGP, the Shaffer Lab is collaborating with Ian Wang’s group at UC Berkeley and multiple state and federal agency scientists to produce landscape genomic data and analyses for 34 species of reptiles and amphibians covering the breadth of California. Our goal is to produce a comprehensive analysis of the geography of genetic variation to guide management and habitat protection now and in the face of climate change.
California Tiger Salamander
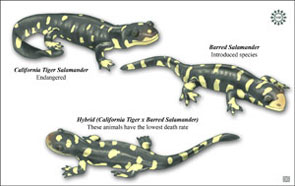 A primary focus of our lab has always been the federally and state listed California tiger salamander, Ambystoma californiense. The species has declined across its entire range, which spans central California from Sonoma to Santa Barbara Counties. Our work has provided the scientific backing both for listing the species, and for delimiting patterns of gene flow and population structure that are central to our ecological, evolutionary, and conservation-oriented work. Most recently, our work has focused on two topics. The first is an amazing case of hybridization between non-native, introduced Barred tiger salamanders and native California tiger salamanders. It’s a story that has as much to say about mechanisms of speciation as it does about conservation biology, and we continue to explore the system from a variety of genetic, genomic, and ecological perspectives. The second is the conservation and recovery of the critically endangered Santa Barbara Distinct Population Segment, which we have demonstrated as declining to the point that inbreeding depression is occurring in nature. Working with the USFWS, our goal is to turn around this decline and recover the population.
A primary focus of our lab has always been the federally and state listed California tiger salamander, Ambystoma californiense. The species has declined across its entire range, which spans central California from Sonoma to Santa Barbara Counties. Our work has provided the scientific backing both for listing the species, and for delimiting patterns of gene flow and population structure that are central to our ecological, evolutionary, and conservation-oriented work. Most recently, our work has focused on two topics. The first is an amazing case of hybridization between non-native, introduced Barred tiger salamanders and native California tiger salamanders. It’s a story that has as much to say about mechanisms of speciation as it does about conservation biology, and we continue to explore the system from a variety of genetic, genomic, and ecological perspectives. The second is the conservation and recovery of the critically endangered Santa Barbara Distinct Population Segment, which we have demonstrated as declining to the point that inbreeding depression is occurring in nature. Working with the USFWS, our goal is to turn around this decline and recover the population.
Turtles of the World
Turtles are an amazing group- 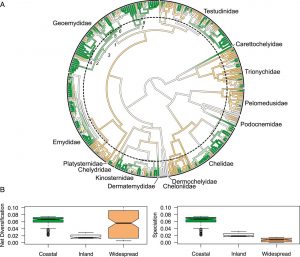 distinctive, easy to wrap your head around, admired and loved by many people, and arguably the most endangered major clade of vertebrates on earth. A central project in the lab has been to acquire 2-3 tissue samples of every species of turtle on earth, sequence them for ~20 independent genes, and build a phylogeny for all 350 or so species of turtles and tortoises that are living today. Our paper, published in PNAS and covered in an article in UCLA Newsroom, reveals new findings about turtles’ evolution so far and their uphill battle for survival.
distinctive, easy to wrap your head around, admired and loved by many people, and arguably the most endangered major clade of vertebrates on earth. A central project in the lab has been to acquire 2-3 tissue samples of every species of turtle on earth, sequence them for ~20 independent genes, and build a phylogeny for all 350 or so species of turtles and tortoises that are living today. Our paper, published in PNAS and covered in an article in UCLA Newsroom, reveals new findings about turtles’ evolution so far and their uphill battle for survival.
Mojave Desert Tortoise
 With funding from the state of California and the U.S. Fish and Wildlife Service, we have developed a radical new genomic toolkit to learn more about how desert tortoises use their habitat and how to avoid conflicts with green energy projects in the Mojave Desert. By using a genomic approach, we have resequenced hundreds geolocated tortoise genomes to learn how they move and what corridors they prefer in key areas of the desert, and how to reduce the impact of alternative placements of solar installations on habitat connectivity from a tortoise perspective (available from bioRxiv here). Our first major publication on the tortoise, published in Science, leveraged this landscape genomic resource and demonstrated that surviving translocation to a new habitat was explained by an individual tortoise’s heterozygosity, rather than its site of origin.
With funding from the state of California and the U.S. Fish and Wildlife Service, we have developed a radical new genomic toolkit to learn more about how desert tortoises use their habitat and how to avoid conflicts with green energy projects in the Mojave Desert. By using a genomic approach, we have resequenced hundreds geolocated tortoise genomes to learn how they move and what corridors they prefer in key areas of the desert, and how to reduce the impact of alternative placements of solar installations on habitat connectivity from a tortoise perspective (available from bioRxiv here). Our first major publication on the tortoise, published in Science, leveraged this landscape genomic resource and demonstrated that surviving translocation to a new habitat was explained by an individual tortoise’s heterozygosity, rather than its site of origin.
CaliPopGen: A genetic and life history database for the fauna and flora of California
What do we really know about the genetics of natural populations of plants, animals, and marine organisms in California? And why should we care? Thousands of papers have been published over the last 40 years using dozens of techniques on hundreds of species, but no synthesis has ever attempted to summarize results from this disparate, scattered set of studies. In a paper published in Scientific Data, we compile, synthesize and analyze this vast literature for the first time. Our goal is to determine how the complex topography and ecology that characterizes California leads to connectivity, isolation, and regional genetic diversity across species and ecosystems. Our results synthesize the published literature on the genetics of natural populations in the Golden State, and answer questions including: are urban populations at greater risk than wildland ones due to inbreeding depression? Are populations of plants and animals on public lands more resilient than those on private lands? What are the major barriers to plant and animal movement across California? The answers to these and many other critical conservation questions are extracted from the published literature by postdocs Joscha Beninde and Erin Toffelmier, faculty lead Brad Shaffer, and a team of nine UCLA undergraduates.
